Advanced imaging techniques provide holistic observations of complicated biological phenomena across multiple scales, while posing great challenges on data analysis. We summarize recent advances and trends in bioimage analysis, discuss current challenges towards better applicability, and envision new possibilities.
“More is different.” As indicated by Philip W. Anderson about 50 years ago, the interactions of large-scale elementary units may cause new properties to emerge, which cannot be explained by the basic laws governing the elementary unit. Biology is a typical discipline exhibiting such a nature of hierarchical structures with heterogenous properties across multiple spatial and temporal scales, e.g., ranging from genes, proteins, organelles, and cells to tissues, organs, and whole bodies. Most of these scales have been covered by advanced instruments such as sequencing technology, cryo-electron microscopy, super-resolution fluorescence microscopy, and magnetic resonance imaging, leading to many discoveries on the molecule and subcellular levels. However, there is a long-standing gap in mesoscale imaging linking cells, tissues, and organs. To fill in the niche, the data throughput of fluorescence microscopy has increased by orders of magnitude in the past decade, opening up a new horizon for investigating large-scale intracellular and intercellular interactions in different pathological or physiological states. Therefore, learning how to process, analyze, and understand large-scale imaging data efficiently to catalyze biological discoveries becomes more and more important. Over the past decade, scientists from different areas have invested collaborative efforts to push forward this frontier for diverse applications such as whole-brain vascular topology, molecular heterogeneity of synaptic morphology, and cell lineages during embryogenesis.
Recent advances and trends. About a decade ago, deep convolutional networks demonstrated their dominance on computer vision for the first time. Since then, deep learning becomes a mainstay in contemporary image analysis due to its key advantages in both efficiency and performance. This revolution rapidly spread to the field of microscopy, and various intelligent methods have been proposed for different biological applications and imaging modalities to solve the problems of image enhancement, classification, segmentation, cell tracking, and others.
Self-supervised and unsupervised learning. For quite a long time, including today, supervised learning is still the primary paradigm of deep-learning-based image analysis. With enough training images and paired ground truth at hand, it is not difficult to construct a model with good performance. However, ground-truth data are not easy to obtain and sometimes even impossible in microscopy, which has become an inevitable limitation of supervised learning. In recent years, the most impressive trend in image analysis is the transition from conventional supervised learning to self-supervised and unsupervised learning. Using these new training mechanisms, networks can learn to perform specific tasks without requiring any paired ground truth for training. Researchers can focus more on designing experiments and exploring biological insights rather than annotating or capturing ground-truth images. Moreover, a typical feature of optical microscopy is the great data variability between different model organisms, experimental conditions and laboratories. It is a common problem that pre-trained models cannot be applied to new data. Self-supervised and unsupervised learning provide a better solution to train a customized model for a specific group of data. For large-scale image analysis, self-supervised and unsupervised methods have an inherent advantage because the raw data itself is a large-scale training set, paving the way for training large models with better generalization.
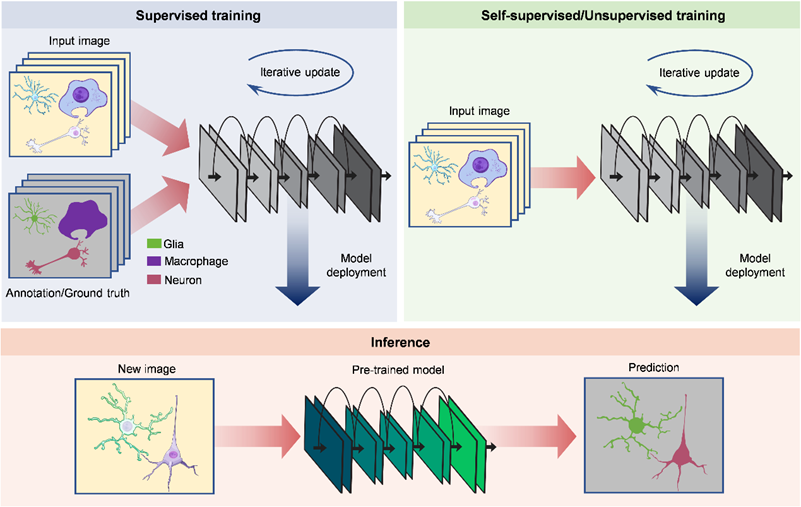
Figure. 1 | Supervised and self-supervised/unsupervised learning for image analysis.
Vision transformers. Network architecture also continues to undergo remarkable updates and to go beyond classical convolutional networks. Vision transformers, a new type of architecture mainly utilizing the self-attention mechanism to extract intrinsic features, have achieved state-of-the-art performance on a variety of computer vision tasks. Their capability to capture long-range dependencies can overcome the locality of convolutional kernels. Just as the success that transformers have attained in predicting genetic variants from long DNA sequences, there is big potential to discover new phenomena by exploiting long-range spatiotemporal correlations in large-scale imaging data. For example, in neural functional imaging, transformers can help to reveal the causality between two distant events in long-term recording and characterize the relationship between two faraway neurons in mesoscale imaging.
Reinforcement learning. As a computational model rooted in the decision-making process of humans and other animals, reinforcement learning is widely used to build intelligent agents with the ability to interact with the environment by rewarding desired behaviors and punishing incorrect ones. Combined with image analysis, reinforcement learning promises to uncover hidden patterns in large-scale imaging data. For example, by treating the migrating cell as an agent and the other cells as the environment, deep reinforcement learning can infer the mechanism of cell migration during embryo development. Reinforcement learning is also suitable for deciphering the neural mechanisms of animal behavior. If we have a large amount of neural imaging data and synchronized visual input and sensory stimuli, it is possible to build cognitive models in a data-driven manner using reinforcement learning. Such a paradigm can be extended to diverse fields to extract huge amounts of biological discoveries through the screening of large-scale imaging data during different pathological or physiological states in different organs. Another success of deep reinforcement learning is to train intelligent agents to complete various challenging tasks. This inspires us that, by designing the reward properly and learning from a large amount of experiment data iteratively, a microscope can learn to interact with the sample, complete a given imaging task with optimal parameters, and discover interesting phenomena all automatically.
Physics-informed deep learning. Imaging is a rigorous optical process that needs to be reliable enough to support scientific discoveries. The analysis of imaging data, especially image processing, should maintain this reliability as much as possible. For low-level vision tasks aimed at improving image quality (e.g., denoising, super-resolution reconstruction, deblurring, etc.), incorporating the physics of image formation into the processing framework can increase the confidence level of the results. Moreover, the difference in physical models of different imaging modalities is also worth considering. In fact, optical imaging is a means of quantitative measurement. The intensity of each pixel has a specific biophysical and biochemical meaning, such as ion concentration in calcium imaging, membrane potential in voltage imaging, and gene expression in spatial transcriptomics. Preserving the quantitative property in processed images is therefore critical for researchers to decode underlying biological phenomena.
Real-time processing. While post-processing is still the main form of image analysis so far, some recent work has begun to integrate the processing tool into the data acquisition pipeline to circumvent the considerable time consumption required for large-scale image analysis. Real-time processing allows researchers to inspect results immediately after capturing raw data, which makes it possible to adjust experimental strategies during the imaging session. For those applications that necessitate real-time interaction with the sample, such as image-guided single-cell surgery and optogenetic manipulation, post-processing is infeasible and real-time processing cannot be replaced. Implementing real-time processing on an imaging system not only means improving processing throughput, but also requires strict timing alignment and effective synergy between software and hardware. Any method that claims to achieve real-time processing should be experimentally demonstrated on a real system.
Challenges towards better applicability. Although impressive achievements have been made in bioimage analysis, there are still some issues that hinder the applicability of these tools in optical microscopy. Here we summarize several key points and hope that concerted efforts could be paid by the research community to resolve these issues.
Standard and metrics. Publishing standard validation datasets and setting corresponding performance metrics can promote the rigorous development of bioimage analysis methods. Since images from different imaging modalities vary greatly, a specific dataset should be archived for each modality. These validation datasets must be representative enough and cover a wide range of samples and imaging conditions. For intuitive comparison and ranking, the performance metric of each dataset is preferably a number that can reflect the comprehensive capability of a method. Efficiency must be emphasized for large-scale analysis to facilitate practical applications.
Interpretability and reliability. Despite the superior performance, deep learning in microscopy suffers from a trust crisis because of its black-box nature. It is a long-standing challenge to endow deep neural networks with interpretability. Realizing interpretable deep learning requires innovations in the most fundamental concepts. But some technical treatments such as feature visualization and physical-based modeling can improve the reliability to some extent. The biggest concern of most researchers is the potential artifacts in the results. It is beneficial to relieve the trust crisis if we have practical approaches to evaluate these errors quantitatively. For example, in addition to the results, giving the corresponding confidence levels at the same time is helpful.
General platform. State-of-the-art image analysis methods are built on the latest advances in computer vision. Using these methods requires strong programming skills and professional background, which can be troublesome for biologists without computational expertise. The most successful platform for biological-image analysis is Fiji based on ImageJ. It integrates abundant classical processing methods in an interactive manner. However, Fiji has lagged behind the recent prosperity of deep learning. There is an urgent demand for a new interactive platform to implement deep-learning-based image analysis. A good platform should not only enable the deployment of pre-trained models, but more importantly, support the training of new models. These operations are all computationally demanding, so the platform must consider how to have easy access to local or cloud-based computing power. Since most current deep learning methods are based on Python, the new platform should be compatible with Python to make full use of open-source resources.
Data sharing. The huge amount of data in biological imaging makes data sharing quite difficult. To facilitate the sharing of large-scale datasets, firstly, web platforms supporting online preview and download of high-dimensional imaging data are needed. Secondly, high-efficiency lossless compression should be applied to reduce the requirements on transmission bandwidth and storage devices. Moreover, a new data format that divides the original large-scale dataset into many units would be useful, as each unit is independently available and meaningful. The metadata should include not only the basic description of the dataset but also thumbnails of the dataset. Users can then have a concrete understanding without downloading the whole dataset.
New possibilities offered by emerging technologies. Recent research in artificial intelligence (AI) suggests that large language models (LLMs) can have human-level performance in language comprehension, reasoning, and programming. Among them, ChatGPT and multimodal GPT-4 have aroused wide attention owing to their ability to understand users and respond fluently on various topics. The great potential of LLMs provides the possibility to build a professional model to help us analyze imaging data. This is an AI engineer that can understand our needs and process the data automatically, such as writing custom scripts and training specific deep learning models. All we need to do is to upload our data, enter our requirements in the dialog box, and give some intermediate comments, which effectively relieves the heavy burden of large-scale image analysis. However, there is still a long way to go to accumulate enough training examples and train such an intelligent and professional model. Open access to code and data of published papers is critical to realize this long-term goal. It is worth noting that AI can only do repetitive work and pioneering contributions still need to be made by humans. Additionally, a widespread concern is that current LLMs sometimes give fictitious answers39. Strict criteria and validations must be in place to ensure they are properly used in image analysis.
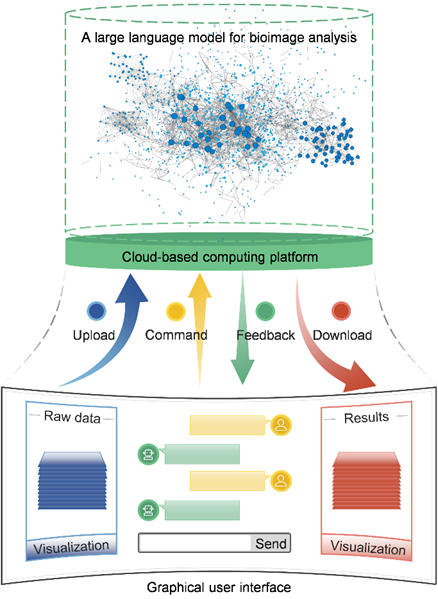
Figure. 2 | Using Large language models for analyzing bioimaging data.
The growing demand for large-scale image analysis poses another great challenge on computing power. Conventional silicon processors can hardly satisfy the requirement for high-speed processing. Optical computing is an emerging technology that processes information at the speed of light by using photons instead of electrons for computation. Not only the inference of deep neural networks, but also basic matrix operations could be implemented by integrated photonic circuits. The results of image analysis could be obtained instantaneously with the acceleration of photonic chips. Combining image analysis with optical computing promises to improve the processing speed to a much higher level and enable data-heavy and high-throughput applications such as single-cell sequencing and image-based high-content screening.
Paper link: https://www.nature.com/articles/s43588-023-00568-2