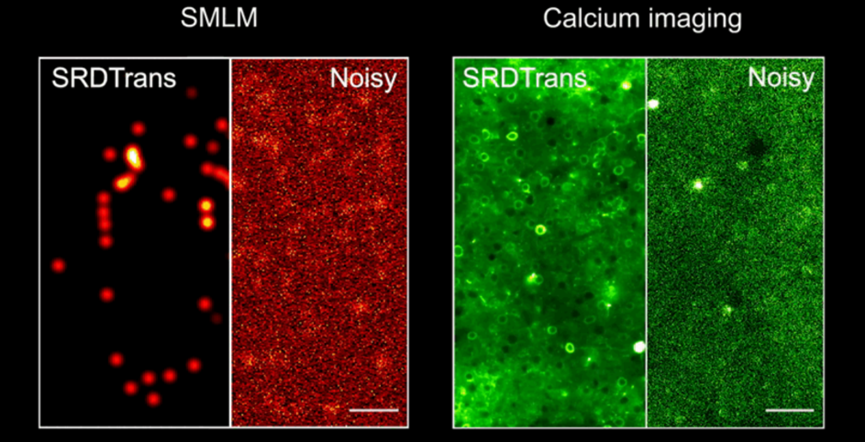
The rapid development of optical imaging technology has enabled researchers to observe the structure and activity of organisms on a micron and even nanometer scale. Fluorescence microscopy, a widely used imaging method, aids in discovering new physiological and pathological mechanisms due to its high spatiotemporal resolution and molecular specificity. The primary goal of fluorescence microscopy is to obtain high signal-to-noise ratio (SNR) images containing sufficient sample information for accurate downstream analysis and convincing conclusions. However, limitations such as labeling concentration, probe brightness, phototoxicity, photobleaching, and photon noise make it challenging to achieve high SNR images, especially at low illumination intensity and high imaging speed.
New Sampling Strategy and New Network Architecture
To address this long-standing issue, the article introduces a self-supervised denoising method using a visual self-attention model leveraging spatial redundancy, named SRDTrans. By designing a three-dimensional orthogonal sampling strategy, this method eliminates the dependence on temporal redundancy, excelling in high-speed dynamic scenarios and low imaging speeds. Compared to previous methods, SRDTrans reduces the required imaging speed by two orders of magnitude. Additionally, a lightweight spatiotemporal fusion self-attention model is designed to fully utilize the long-distance correlation in data, enhancing performance and processing speed. SRDTrans offers superior global perception and high-frequency restoration capabilities compared to classical convolutional networks, recovering fine spatiotemporal activities undetectable by other methods and achieving significant performance improvements.
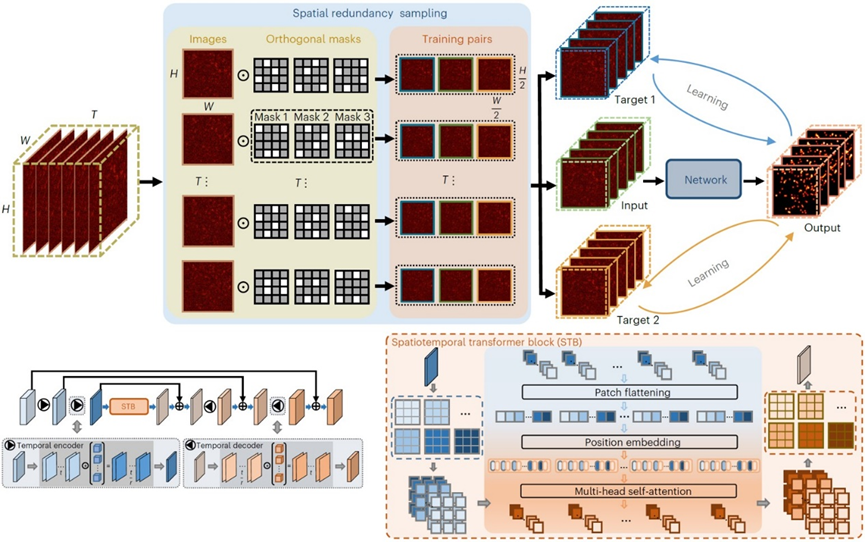
Figure 1. Spatial Redundancy Sampling Strategy and Lightweight Visual Self-Attention Network Architecture
High-performance Single-Molecule Localization Microscopy (SMLM) with SRDTrans denoising
SRDTrans is first applied to single-molecule localization microscopy (SMLM), which captures images of randomly blinking fluorescent molecules with no temporal correlation between adjacent frames. Experimental results show that SRDTrans significantly improves the SNR of denoised data, clearly restoring single-molecule emission patterns drowned by noise, with an average SNR gain of over 20dB. The noise removal greatly enhances downstream single-molecule localization and super-resolution reconstruction, reducing false positives and false negatives, and achieving higher resolution in reconstructed subcellular structures. SRDTrans also performs well on medium-to-high SNR data collected from other laboratories, enhancing microtubule structure localization accuracy and tripling reconstruction resolution.
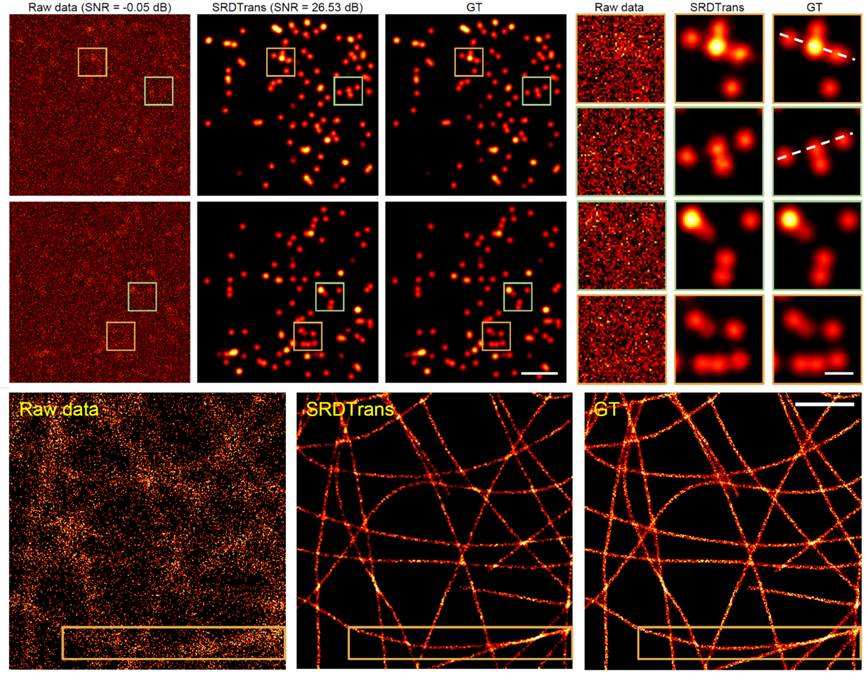
Figure 2. SRDTrans Achieves Ultra-Sensitive Single-Molecule Localization Imaging
SRDTrans Improves Volumetric Multi-Photon Imaging
In multi-photon imaging, the three-dimensional imaging speed linearly decreases with the number of scanning planes, resulting in low 3D sampling rates for large-scale neural activity observation. Temporal redundancy-based denoising methods are thus impractical, but SRDTrans provides a new approach for enhancing weak fluorescence signals in 3D calcium imaging. Experiments show that SRDTrans maintains optimal denoising performance at low calcium imaging speeds of 0.3Hz, without causing fluorescence signal broadening and distortion. This work achieves 3D calcium imaging data denoising for the first time, clearly revealing neuronal spatial contours and firing states in the 500×500×200 μm³ brain tissue of the mouse cortex at 0.3Hz two-photon calcium imaging, avoiding noise interference in neuronal signal extraction. Given the three-dimensional distribution of neural circuits in the mammalian brain, high SNR 3D imaging is essential for analyzing large-scale neuronal networks' functionality. SRDTrans’s superior performance offers new tools for achieving ultrasensitive neural recordings.
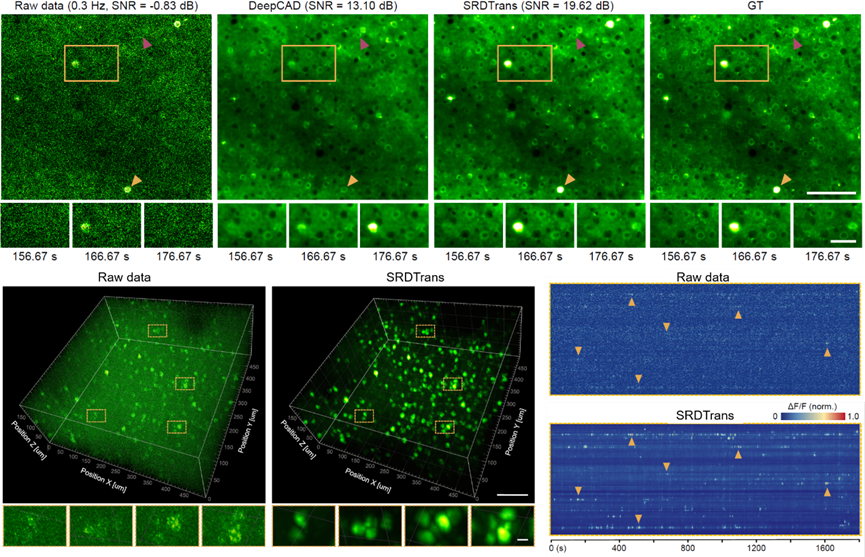
Figure 3. SRDTrans Achieves Ultra-Sensitive 3D Two-Photon Calcium Imaging
SRDTrans Has Good Potential for Fast Dynamic Life Activity Research
With fluorescent probes developing toward faster dynamics to monitor millisecond-level life activities, achieving sufficient imaging speed becomes increasingly challenging. SRDTrans, by overcoming the dependence on high imaging speed, will play a crucial role in studying high-speed dynamic life activities. Furthermore, SRDTrans is not reliant on specific imaging mechanisms, noise models, or sample morphologies, making it easily extendable to other biological samples and imaging modalities such as voltage imaging, wide-field imaging, light-sheet microscopy, confocal microscopy, light-field microscopy, and super-resolution microscopy.
Paper link:https://www.nature.com/articles/s43588-023-00568-2
Code link:https://github.com/cabooster/SRDTrans