With the development of various animal models in recent years, the demand for observing cell-cell and intracellular interactions in vivo has rapidly increased. Due to the complexity of the three-dimensional microenvironment, cells and organelles exhibit markedly different behaviors in cell culture dishes and multicellular organisms. More importantly, their functions and behaviors often show rapid changes over long time scales in different physiological or pathological states. However, imaging of live tissues involves challenges such as light scattering, optical aberrations, sample three-dimensional movement, and phototoxicity, greatly reducing the imaging performance of current microscopy instruments in complex environments (especially within mammalian bodies). The rapid construction of advanced microscopy imaging systems has long been a challenge for many biological laboratories.
On June 29, 2022, the team led by Prof. Dai Qionghai from the Institute of Brain and Cognitive Sciences and the Department of Automation at Tsinghua University, along with the Tsinghua-IDG/McGovern Institute for Brain Research, published a research paper titled "A practical guide to scanning light-field microscopy with digital adaptive optics" in the journal Nature Protocols. Following their publication in Cell in 2021[1], where they introduced the DAOSLIMIT imaging framework, the team further proposed a comprehensive open-source solution for scanning light field microscopy. This solution, based on commercially available optoelectronic devices, serves as an extension module for conventional wide-field fluorescence microscopes. It provides a fully open-source hardware, software, and graphical user interface workflow for implementing scanning light field microscopy, and is also applicable to other types of light field microscopy instruments. This paper represents the first guide to high-resolution light field microscopy technology internationally, offering support for building simple and practical advanced microscopy imaging systems in biological laboratories.

Unlike traditional live microscopy techniques where spatial resolution and angular resolution constrain each other, scanning light field microscopy images the entire volume in a tomographic manner, providing a compact computational solution. As shown in Figure 1, it significantly improves spatiotemporal resolution by synchronizing high-speed scanning mirrors with carefully designed reconstruction algorithms, thereby enhancing imaging efficiency and effectively controlling phototoxicity to live tissues. Additionally, leveraging high-resolution multidimensional spatial angle information, this method decouples signal acquisition from adaptive wavefront correction by acquiring four-dimensional full photon information, digitally estimating and correcting spatially non-uniform aberrations without requiring additional wavefront sensors and spatial light modulators. The digital adaptive optics framework helps ASLFM maintain high resolution and signal-to-noise ratio in complex environments and exhibits strong robustness against system aberrations caused by optical misalignment. Therefore, scanning light field microscopy can achieve high-speed subcellular resolution observation within live tissues while having extremely low phototoxicity, surpassing the conventional confocal microscopy by more than three orders of magnitude.
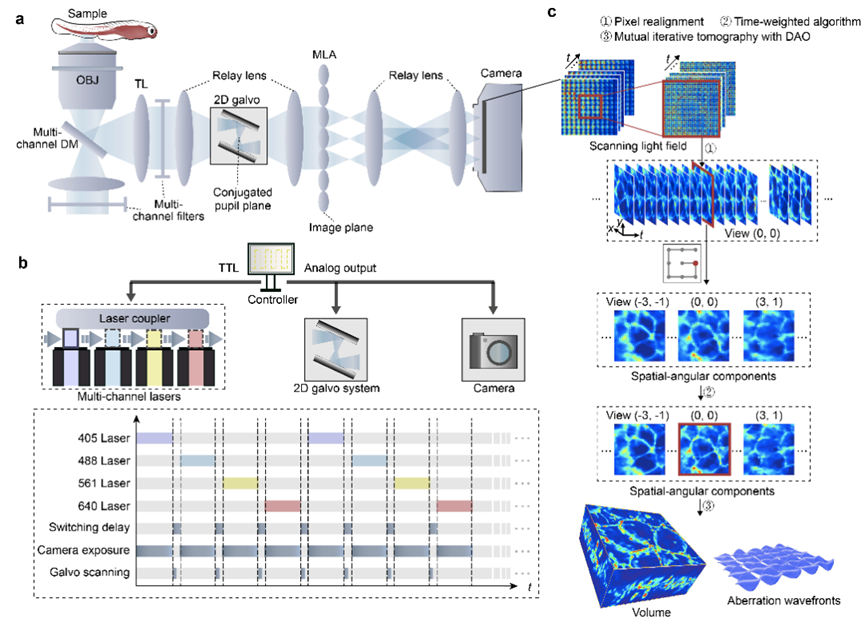
Figure 1. Principle of Scanning Light Field Microscopy
As a computational imaging framework, software reconstruction and hardware system are equally important. When the reconstruction parameters do not match the image acquisition process, artifacts may appear in scanning light field microscopy. Effective artifact diagnosis and accurate parameter selection to compensate for system errors are crucial for scanning light field microscopy. This solution provides a detailed description of potential imaging artifacts of different types and various parameters for correction and performance optimization, as shown in Figure 2. Artifacts such as "stripes", "checkerboard patterns", and "honeycomb patterns", among others, can be systematically removed. Furthermore, to enhance tolerance for parameters such as optical system misalignment and equipment manufacturing differences, this solution offers a one-click calibration procedure, enabling robust imaging performance up to the diffraction limit within live tissues. It also provides an open-source graphical user interface for hardware synchronization control and real-time rendering of multi-angle images.
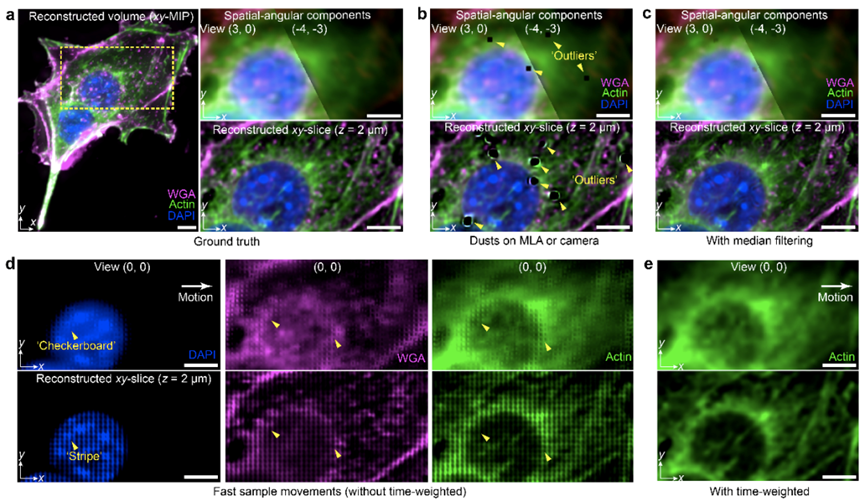
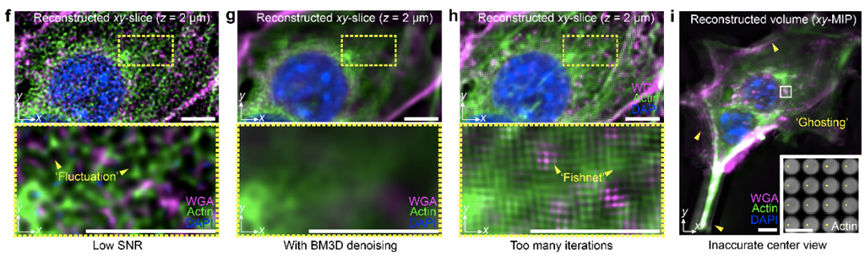
Figure 2. Detailed Analysis and Solution for Artifacts
Finally, the authors demonstrated the spatial resolution of scanning light field microscopy and typical applications in different species, as shown in Figure 3. In complex scenarios such as multi-colored labeled B16F10 cells, migrating neutrophils in live mouse liver, and developing zebrafish embryos, high-resolution live three-dimensional imaging performance was achieved, capturing rapid contraction processes of filamentous pseudopods and dynamic changes of organelles. The high spatiotemporal resolution, digital adaptive correction capability, and low phototoxicity of scanning light field microscopy open new doors for observing interactions between mammalian cells and intracellularly in live organisms. This solution will help researchers achieve advanced imaging performance through low-cost and compact optical systems and can serve as a universal light field imaging framework, further supporting research on complex cell behaviors in different pathological or physiological states within live organisms. The paper includes a complete set of optical path design and mechanical design architectures, along with corresponding commercially available lens selection references. All software and hardware controls and reconstruction algorithms are integrated into a single graphical interface, with interfaces available for further modification, providing a research platform for other light field microscope developers.
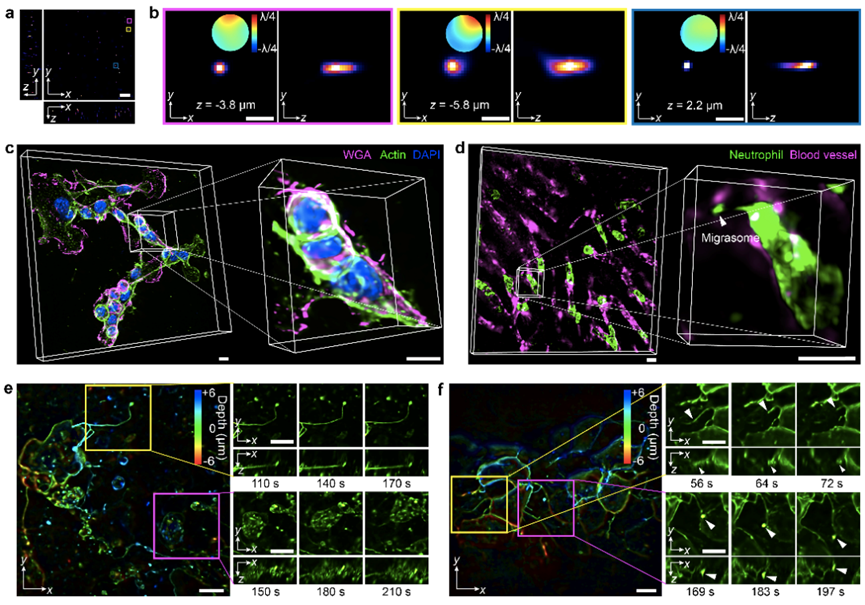
Figure 3. Typical Imaging Cases on Different Biological Samples
The first author of this paper is Lu Zhi, a doctoral student from the Department of Automation at Tsinghua University. Cai Yeyi, Nie Yixin, and undergraduate student Yang Yuxin, all from the Department of Automation at Tsinghua University, contributed to the research. The co-corresponding authors of the paper are Professor Dai Qionghai from the Department of Automation, Beijing National Research Center for Information Science and Technology, Institute of Brain and Cognitive Sciences, and Tsinghua-IDG/McGovern Institute for Brain Research, and Assistant Professor Wu Jiamin.
[1] Wu, J. et al. Iterative tomography with digital adaptive optics permits hour-long intravital observation of 3D subcellular dynamics at millisecond scale. Cell 184, 3318-3332 (2021).
[2] Lu, Z. et al. A practical guide to scanning light-field microscopy with digital adaptive optics. Nature Protocols (2022).