https://github.com/GuoxunZhang-PhD/DeepSeMi
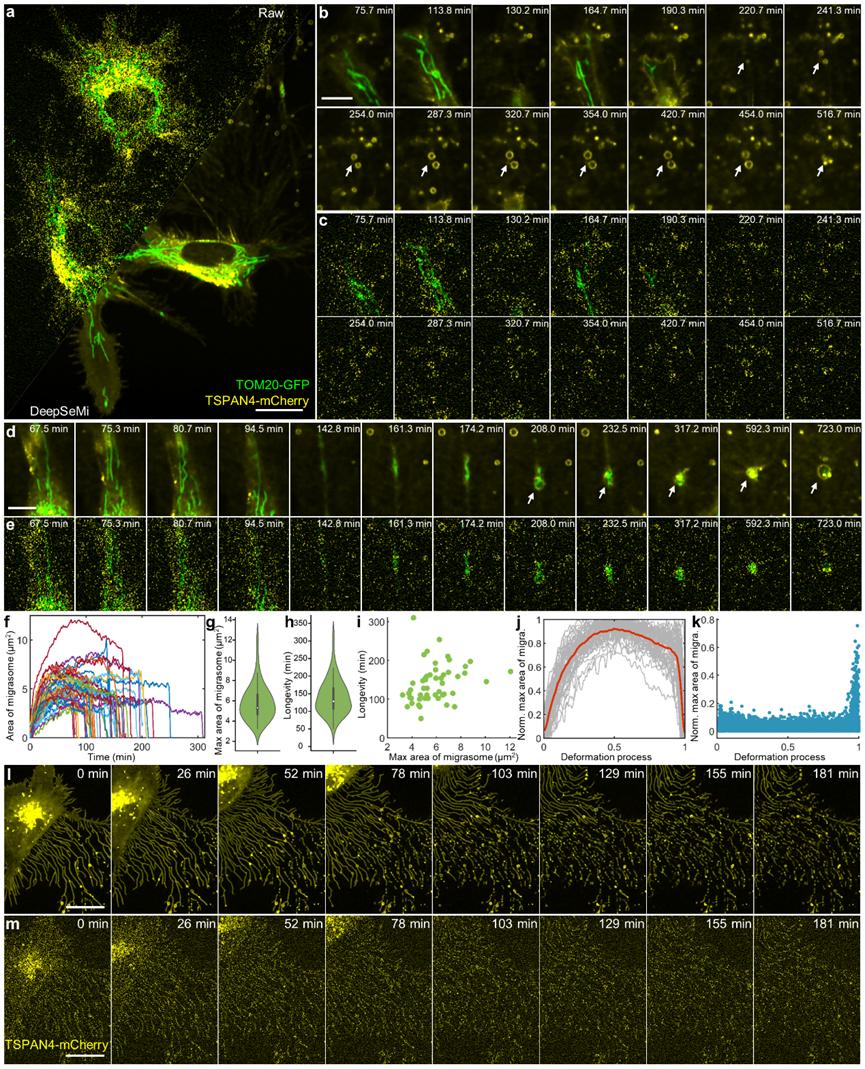
Fluorescence microscopy has become an indispensable tool for revealing the dynamic regulation of cells and organelles. However, stochastic noise inherently restricts optical interrogation quality and exacerbates observation fidelity when balancing the joint demands of high frame rate, long-term recording and low phototoxicity. Here we propose DeepSeMi, a self-supervised-learning-based denoising framework capable of increasing signal-to-noise ratio by over 12 dB across various conditions. With the introduction of newly designed eccentric blind-spot convolution filters, DeepSeMi effectively denoises images with no loss of spatiotemporal resolution. In combination with confocal microscopy, DeepSeMi allows for recording organelle interactions in four colors at high frame rates across tens of thousands of frames, monitoring migrasomes and retractosomes over a half day, and imaging ultra-phototoxicity-sensitive Dictyostelium cells over thousands of frames. Through comprehensive validations across various samples and instruments, we prove DeepSeMi to be a versatile and biocompatible tool for breaking the shot-noise limit.
DeepSeMi enables half-day-long observations of migrasomes and retractosomes with low phototoxicity.
Zhang, G., Li, X., Zhang, Y. et al. Bio-friendly long-term subcellular dynamic recording by self-supervised image enhancement microscopy. Nat Methods 20, 1957–1970 (2023). https://doi.org/10.1038/s41592-023-02058-9