Whole Brain Cell Atlas of Mice
Introduction
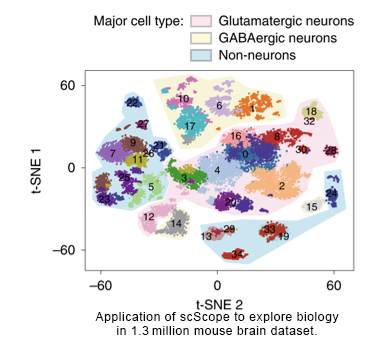
Recent advances in large-scale single-cell RNA-seq enable fine-grained characterization of phenotypically distinct cellular states in heterogeneous tissues. We present scScope, a scalable deep-learning-based approach that can accurately and rapidly identify cell-type composition from millions of noisy single-cell gene-expression profiles.
Highlights
Scalable analysis of cell-type composition
Recursive self-coding neural network
Large-scale fast clustering based on down-sampling, K-means clustering and graph clustering
Rapid analysis of mega-scale intracellular structures
Deng, Y., Bao, F., Dai, Q., Wu, L. F., & Altschuler, S. J. (2019). Scalable analysis of cell-type composition from single-cell transcriptomics using deep recurrent learning. Nature methods, 1.